Setup ABIDE Preprocessing
Preprocessing
Overview
The redoing of the ABIDE preprocessing will center around the use of ANTS instead of FNIRT for the non-linear registration. There are a couple of setup points:
- Do eigenvector separately since it takes so long
- Make sure to run only a certain set of subjects at once on gelert
- If I set the working directory to /tmp whilst using gelert, then this could speed up preprocessing.
I’m using a version of CPAC with the following path: /home/data/Projects/CPAC_Regression_Test/2013-10-04_v0-3-2/run/lib/python2.7/site-packages. This version has the ANTS amongst other things and will be part of the upcoming release. For some reason, I had to re-install nipype. I know there is a 0.9.0 development version of nipype, whereas I install the 0.8.0 stable version. Hopefully, this will be ok.
Setup Config File
The sections below are labeled according to the section in the CPAC GUI edit window. As a summary the following important preprocessing options were selected:
- ANTs for anatomical registration
- The functional images and their potential derivatives were registered to 3mm standard space.
- Nuisance regression included two strategies that both included motion (Friston 24), linear, and quadratic: * compcor * compcor + global
- Temporal filtering (0.01-0.1 Hz) was applied and not applied
- Spatial smoothing of 4mm was applied to the derivatives.
An issue is that I have set the working directory to be removed after completion. However, this would make it difficult to then run derivates in a subsequent step. Will need to investigate this issue.
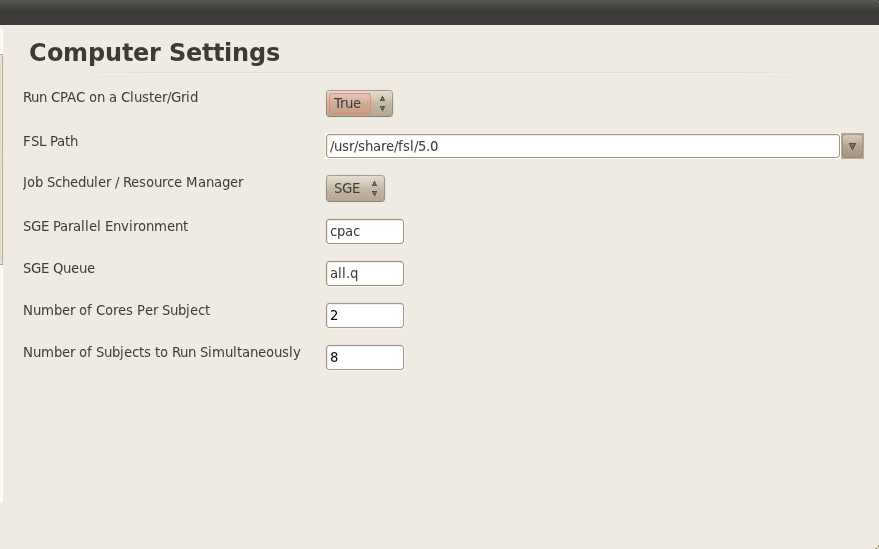
Computer Settings
I have the SGE turned on and will use about 2 cores per subject.
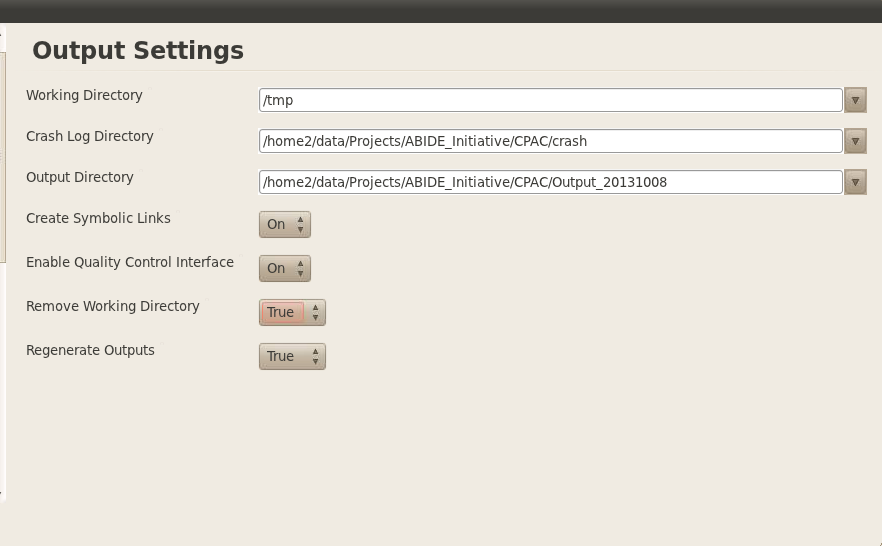
Output Settings
The working directory will be in the tmp directory. Since we are running on the grid engine, this will ensure that the working directory will be local to each node and improve I/O performance. Due to this fact, I have checked removing the working directory.
Preprocessing Workflow Options
Nothing new to see here.
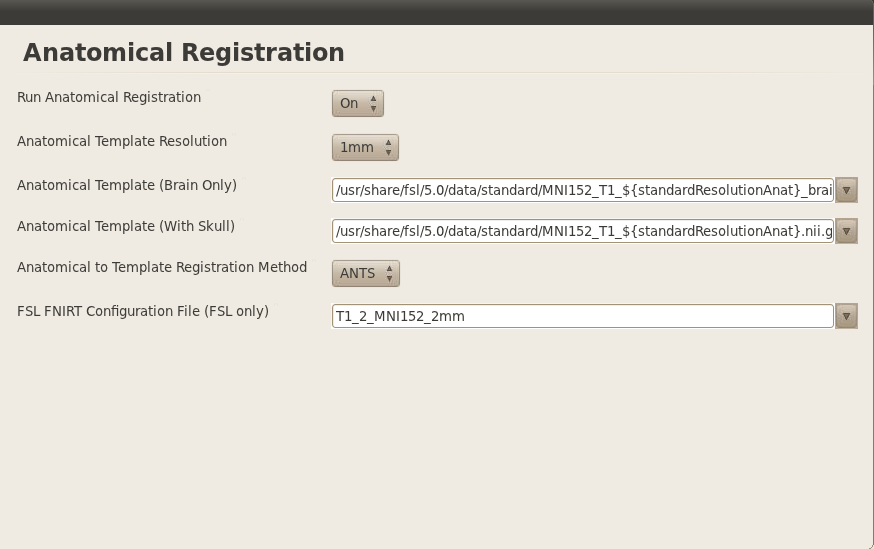
Anatomical Preprocessing
We are running ANTs for the registration, otherwise everything else is standard.
Functional Preprocessing
Time Series Options
Left this to be the standard. I’m assuming that since there are multiple sites, I’ll have the first timepoint and the TR in a csv type file.
Functional to Anatomical Registration
This seems to have a Functional Standard Resolution as an option but it should actually be in the following Functional to MNI section, no? Besides this blemish, no changes were made from the default.
Nuisance
I did not do median angle correction, but I did set to types of nuisance corrections:
- compcor, motion, linear, and quadratic
- same as number 1 and global
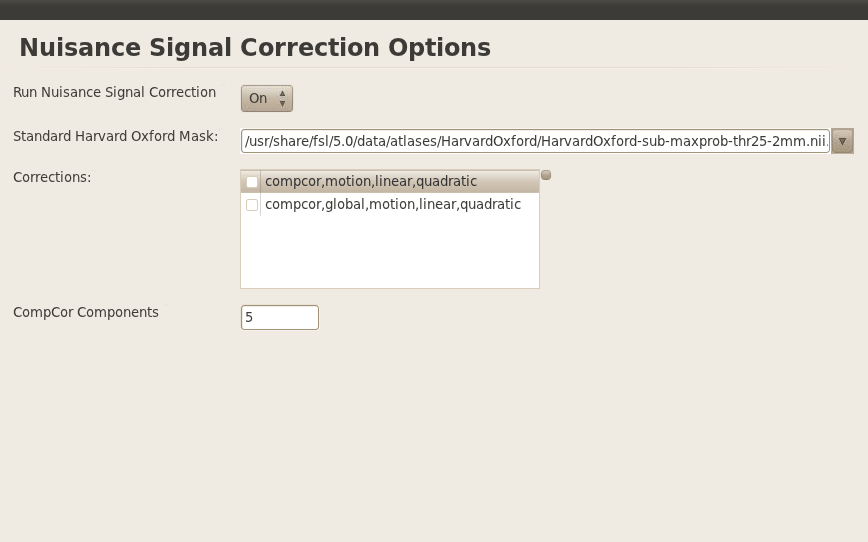
Please note that in the above image, I didn’t select the two correction (checkbox) but I did so later on.
Temporal Filtering Options
On the GUI, I set this to just ‘On’ but later I in the yaml config file I will set temporal filtering as both on and off. The band-pass filtering is set to 0.01-0.1 Hz.
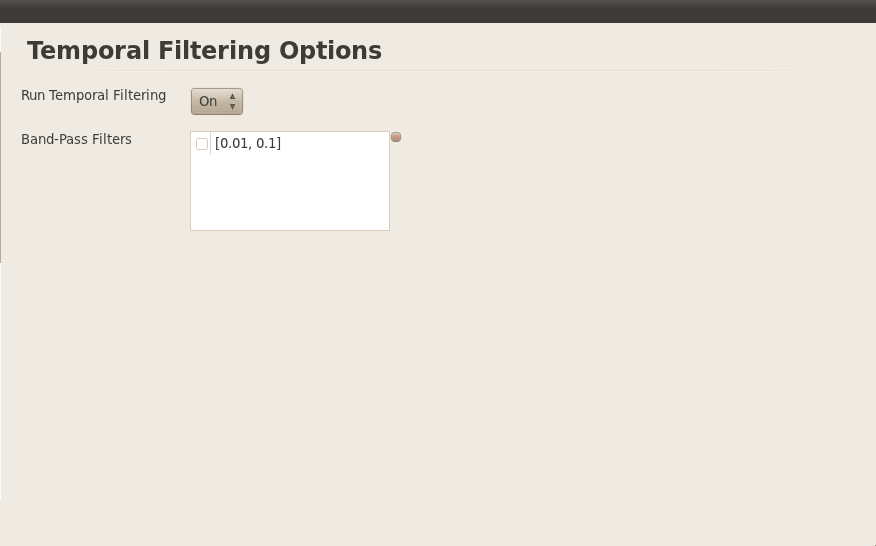
Same thing with the temporal filtering, the box should by checked next to the bandpass thingy.
Motion Correction
This is set to on and I’m using the Friston 24 parameter model. Scrubbing is turned off (no scrub here).
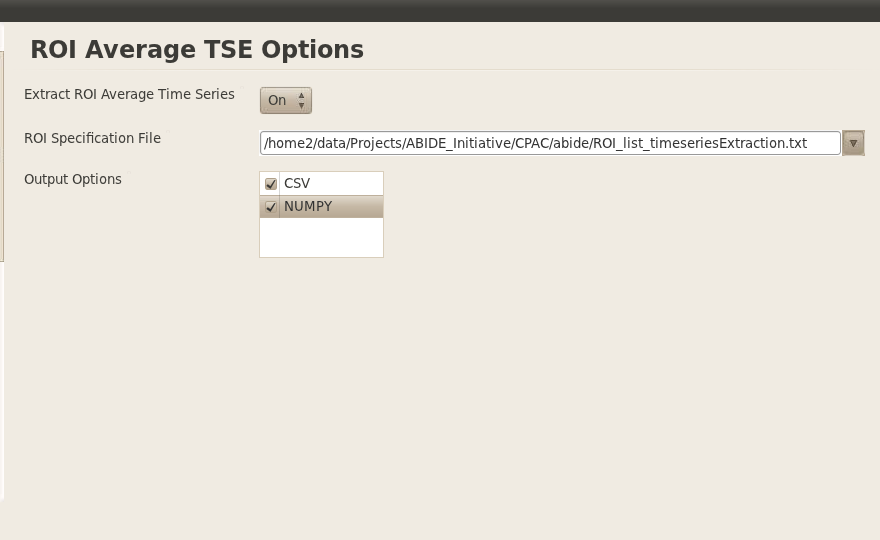
Time Series Extraction (TSE)
The creation of the ROIs from the coordinates in Dosenbach et al., 2010 paper with 160 ROIs has already been done. So I won’t redo it here. However, if one did want to regenerate those ROIs, then you would need /home2/data/Projects/ABIDE_Initiative/CPAC/abide/Dosenbach_Science_160ROIs.txt as the specification file and give your own output directory.
The time series was extracted from several ROIs. These include various anatomical and functional parcellations as well as the regions-of-interest (160) from the Dosenbach et al., 2010 Science paper. The nifti files to these different parcellations and ROIs are also provided on the github repository in the templates/3mm folder.
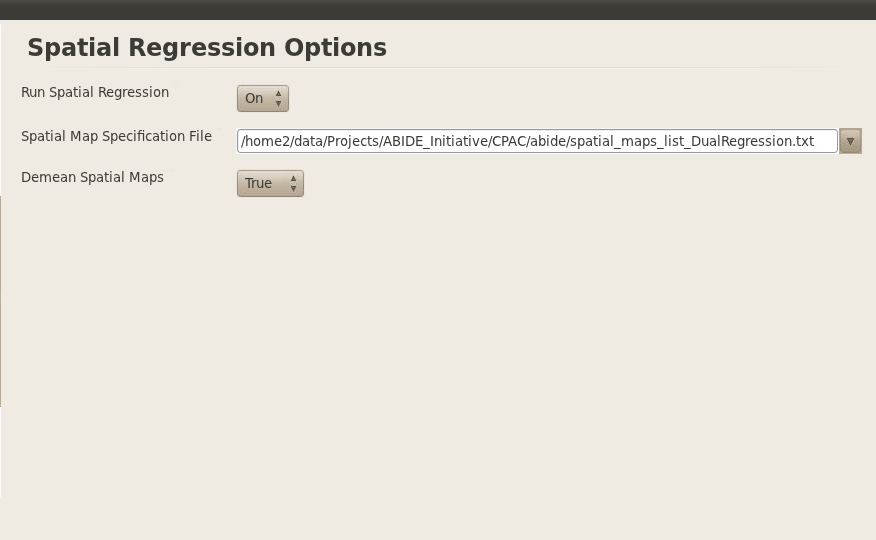
Finally, I set spatial regression to a yes and we used spatial maps from the ICA in the PNAS Smith et al., 2009 paper.
Spatial Smoothing
This has been left at the default of 4mm.
Other Derivatives
Note that for this stage, I haven’t selected any of the derivatives. Those will be set in another config file. For the centrality derivatives, I will also need to create a grey matter image from all of the subject’s data.